RNA Clustering using Graph Neural Networks Embeddings
You can find attach the report and code. Work done with Yann Ponty, Sebastian Will and Johannes Lutzeyer.\
RNA are essential molecules in the cell, playing a key role in the translation of the genetic code into proteins. They can fold into complex structures that are generally modelled as planar graphs, a graph where the nodes are connected by edges that do not cross each other, called the secondary structure.\
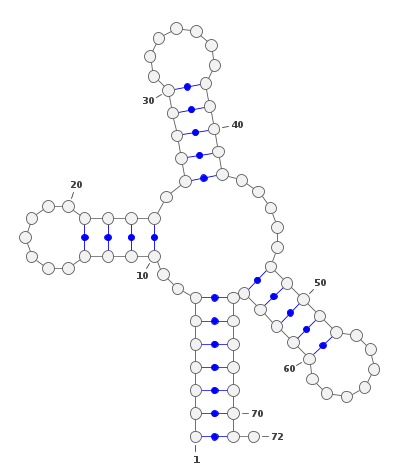
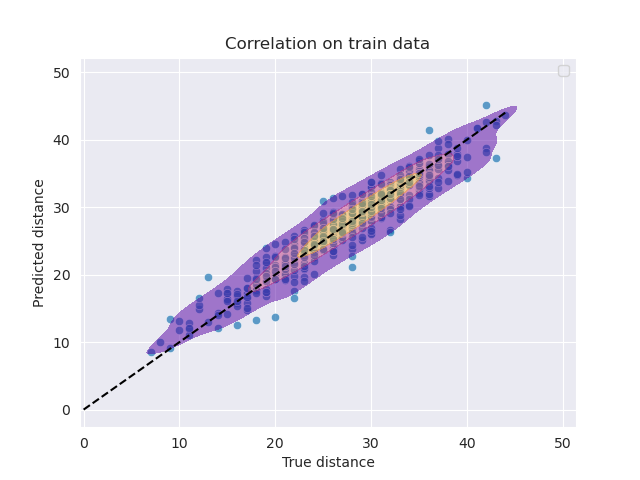
For bioinformatician, analysis of large RNA databases is a common task, with the goal of clustering similar RNA structures to find different conformations of the same RNA. A K-Means Clustering using a precomputed distance, the base-pair distance (Symetic difference of the set of edges). This is a computationally expensive task as the distance matrix is of size $O(n^2)$ where $n$ is the number of RNA.\
A strategy is to obtain a low-dimensional representation of the RNA secondary structure that captures the essential information for clustering using a GNN. K-Means clustering is then linear in the number of RNA.\
PyTorch Geometric is used to implement and train the GNN. The model used is a Residual Gated Graph Convolutional Network with a distance loss:
\[\mathcal{L}_{distanceLoss} = (\Delta(E_1, E_2) - ||\mathbf{X}_{1,pool} - \mathbf{X}_{2, pool} ||)^2\]where $\Delta(E_1, E_2)$ is the base-pair distance between the two RNA, $\mathbf{X}{1,pool}$ and $\mathbf{X}{2, pool}$ are the pooled embeddings of the two RNA.\